R interface (rBExIS)
For new rBExIS version (updated on 2023-03-13):
see: https://github.com/BEXIS2/rBExIS#preparation
Requirements
- R (e.g. https://cran.r-project.org/bin/windows/base/ for Windows, otherwise start here)
- RStudio (https://www.rstudio.com/products/rstudio/download/#download)
- Rtools (e.g. https://cran.r-project.org/bin/windows/Rtools/ for Windows, , otherwise start here)
Installation
To uninstall the current version:
detach("package:rBExIS", unload = TRUE)
remove.packages("rBExIS")
To install the new version:
The script depends on the packages httr, jsonlite, XML, rBExIs, and tidyverse. rBExIs installation also depends on devtools. If they are not yet installed, they will be installed in the next chunk of code.
sapply(c("httr","jsonlite","XML","tidyverse","devtools"),
function(x){
if(!require(x,character.only = T)){
install.packages(x)
require(x,character.only = T)
}
})if(!require("rBExIS")) install_github("BEXIS2/rBExIS", subdir = "rBExIS")
require(rBExIS)Database webpage and authorization indification:
bexis.options("base_url" = "https://jexis.idiv.de/")
Within BEXIS2, the functions are either publically available (i.e. no authentication/authorization is needed to execute the function). On the other side, other functions may need authentication to verify the user of that function.
Here it uses the bearer token for authentication.
To get your token number, to go JEXIS page and log in, under your name, click “Token”.
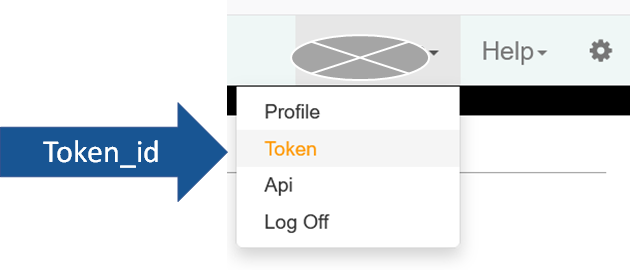
bexis.options("authorization_bearer" = "KJkSKJxEoiXwk9ipAvKkNEJ9isGGi64drtQDRf9KKCDRKSE5JYvz5j8Yx5Unvto5")
Functions
List of Dataset Ids
bexis_dataset_ids <- rBExIS::bexis.GetDatasetIds()
Data of specific Dataset
That function uses the parameter id to identify the specific dataset you would like to get the data from.
bexis_data <- rBExIS::bexis.GetDatasetById(id)
List of Metadata Objects
bexis_metadata_list <- rBExIS::bexis.GetMetadata()
Metadata of specific Dataset
That function uses the parameter id to identify the specific dataset you would like to get the metadata from.
bexis_metadata <- rBExIS::bexis.GetMetadataById(id)
List of Data Structures
bexis_structures <- rBExIS::bexis.GetDataStructures()
Data Structure of specific Dataset
That function uses the parameter id to identify the specific data structure you would like to get the information/details from.
bexis_structures <- rBExIS::bexis.GetDataStructureBy()
For old rBExIS version:
We need the packages httr, jsonlite, XML, rBExIs. rBExIs installation also depends on devtools. If they are not yet installed, they will be installed in the next chunk of code.
sapply(c("httr","jsonlite","XML","devtools"),
function(x){
if(!require(x,character.only = T)){
install.packages(x)
require(x,character.only = T)
}
})
if(!require("rBExIS")) install_github("BEXIS2/rBExIS", subdir = "rBExIS")
require(rBExIS)bexis.options() # info of the url and token number
bexis.options(base_url = "https://jexis.idiv.de/")
token_id<-”XXXXXXXXXX”
bexis.options(token = “token_id")
#get list of all dataset ids
bexis_dataset_ids <- bexis.get.datasets()
#get data from bexis from dataset with id = xy
bexis_data90 <- bexis.get.dataset_by(id = 90)
! The NA value in the database will be read as the maximum double value 1.797693e+307 in R ( .Machine$double.xmax/10) if you use rBEXIS package.
For example, here is the code to correct the the biomass “NA” data from 2020:
a<-.Machine$double.xmax/100
d2020$biomass[d2020$biomass >a]<-NA
