New publication from Marr et al. in Scientific Data: LC-MS based plant metabolic profiles of thirteen grassland species grown in diverse neighbourhoods
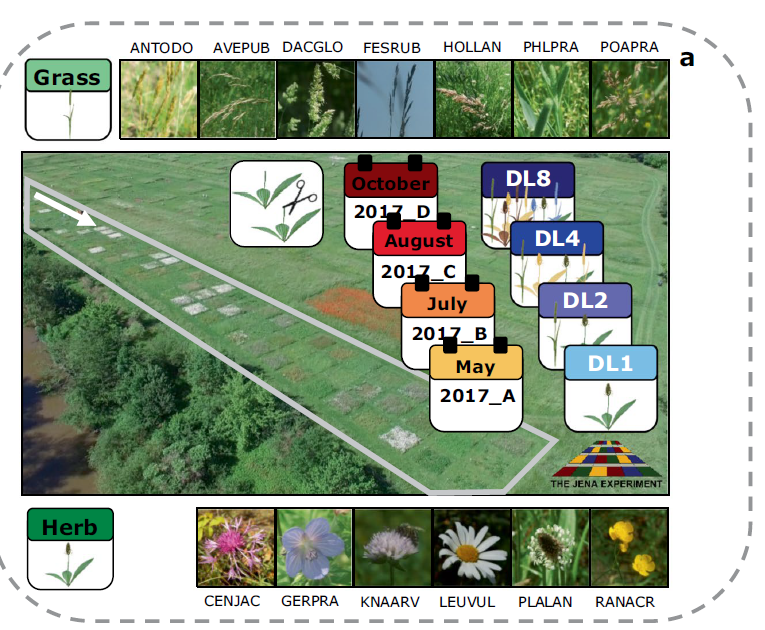
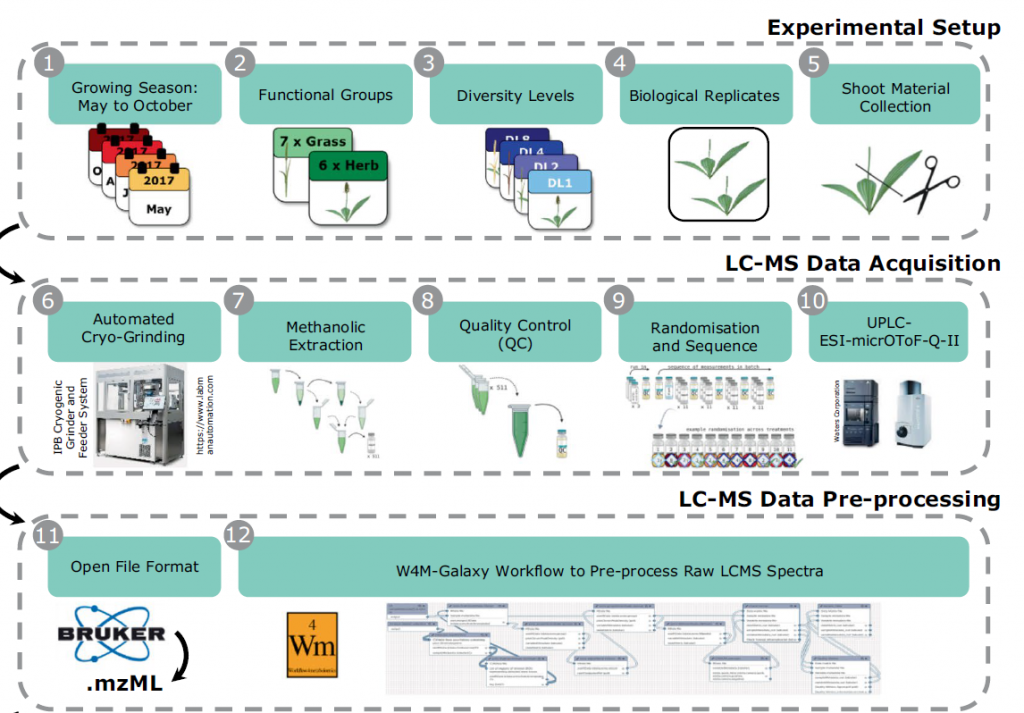
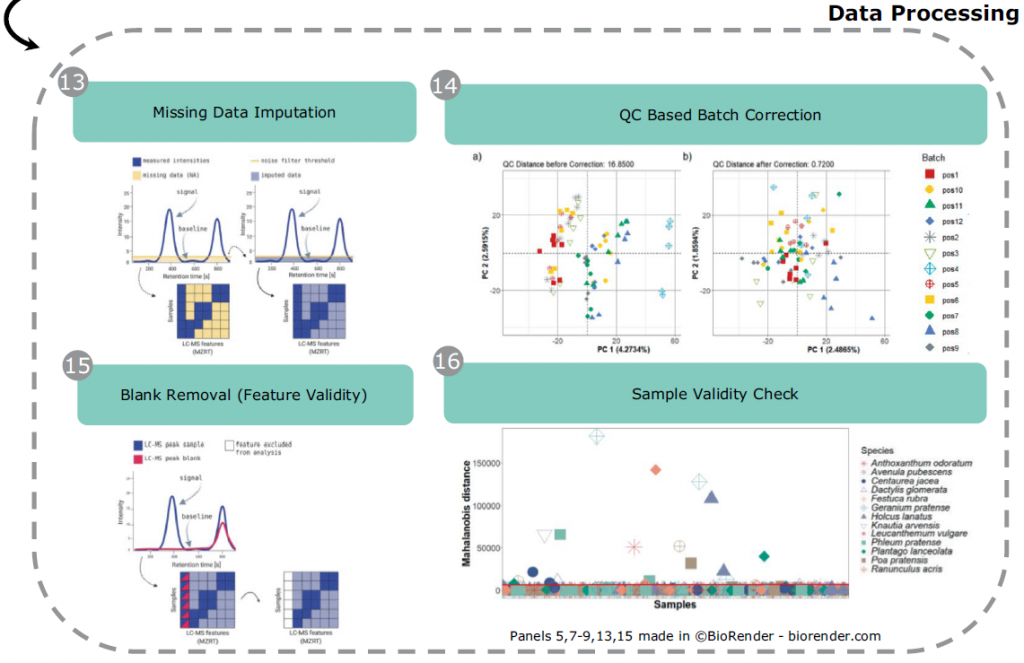
In plants, secondary metabolite profiles provide a unique opportunity to explore seasonal variation and responses to the environment. These include both abiotic and biotic factors. In field experiments, such stress factors occur in combination. This variation alters the plant metabolic profiles in yet uninvestigated ways. This data set contains trait and mass spectrometry data of thirteen grassland species collected at four time points in the growing season in 2017. We collected above-ground vegetative material of seven grass and six herb species that were grown in plant communities with different levels of diversity in the Jena Experiment. For each sample, we recorded visible traits and acquired shoot metabolic profiles on a UPLC-ESI-Qq-TOF-MS. We performed the raw data pre-processing in Galaxy-W4M and prepared the data for statistical analysis in R by applying missing data imputation, batch correction, and validity checks on the features. This comprehensive data set provides the opportunity to investigate environmental dynamics across diverse neighbourhoods that are reflected in the metabolomic profile.
Reference:
Marr, S., Hageman, J.A., Wehrens, R. et al. LC-MS based plant metabolic profiles of thirteen grassland species grown in diverse neighbourhoods. Sci Data 8, 52 (2021). https://doi.org/10.1038/s41597-021-00836-8



